Overlapping genes: Functional characterization
Functional characterization of overlapping genes in bacteria
Group leader until 2021:
PD Dr. Klaus Neuhaus (ZIEL Core Facility Microbiome / NGS)
The coding density of protein coding genes in prokaryotic genomes is probably much higher than previously assumed. Apart from unknown intergenic genes, in some surprising cases the same DNA sequence encodes two proteins in different reading frames (overlapping genes).
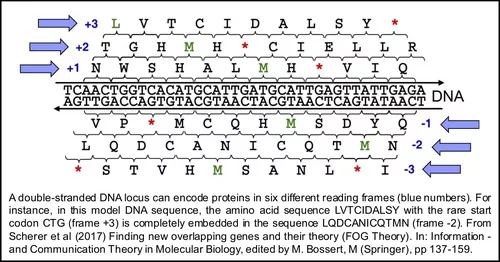
Research on overlapping genes in bacteria is in its infancy and their evolution is an enigma open to scientific investigation. We use experimental approaches to elucidate presence, expression and function of overlapping genes. Such knowledge is important for understanding the de novo origin and evolution of such fascinating gene pairs
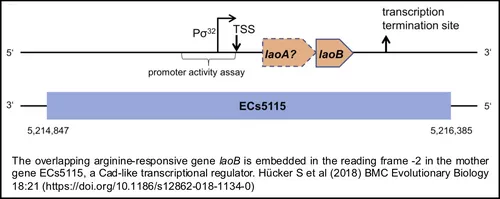
The project started within the Priority Program „Information- and communication theory in molecular biology” (InKoMBio SPP 1395) using an interdisciplinary approach which involved the TUM (Chair of Microbial Ecology, Prof. Siegfried Scherer), Konstanz University (Chair of Data Analysis and Visualization, Prof. Daniel Keim) and Ulm University (Chair “Telecommunications and Applied Information Theory“, Prof. Martin Bossert).
Publications
Zehentner B, Ardern Z, Kreitmeier M, Scherer S and Neuhaus K (2020) A novel pH-regulated, unusual 603 bp overlapping Protein coding gene Pop is encoded antisense to opmA in Escherichia coli O157:H7 (EHEC). Frontiers in Microbiology 11:377. Doi:10.3389/fmicb.2020.00377
Vanderhaeghen S, Barbara Zehentner B, Scherer S, Neuhaus K, Ardern Z (2018) The novel EHEC gene asa overlaps the TEGT transporter gene in antisense and is regulated by NaCl and growth phase. Scientific Reports, www.nature.com/srep/ in press
Hücker SM, Vanderhaeghen S, Abellan-Schneyder I, Wecko R, Simon S, Scherer S , Neuhaus K (2018) A novel short L-arginine responsive protein-coding gene (laoB) antiparallel overlapping to a CadC-like transcriptional regulator in Escherichia coli O157:H7 Sakai originated by overprinting. BMC Evolutionary Biology, BMC Evolutionary Biology 18:21. doi: 10.1186/s12862-018-1134-0
Scherer S, Neuhaus K, Bossert M, Mir K, Keim D, Simon S (2018) Finding new overlapping genes and their theory (FOG Theory). In: Information - and Communication Theory in Molecular Biology, edited by Bossert M (Springer International), pp 137-159.
Neuhaus K, Landstorfer R, Simon S, Schober S, Wright PR, Smith C, Backofen R, Wecko R, Keim DA, Scherer S (2017) Differentiation of ncRNAs from small mRNAs in Escherichia coli O157:H7 EDL933 (EHEC) by combined RNAseq and RIBOseq – ryhB encodes the regulatory RNA RyhB and a peptide, RyhP. BMC Genomics 18:216 DOI 10.1186/s12864-017-3586-9.
Hücker SM, Ardern Z, Goldberg T, Schafferhans A, Bernhofer M, Vestergaard G, Nelson CW, Schloter M, Rost B, Scherer S, Neuhaus K (2017) Discovery of numerous novel small genes in the intergenic regions of the Escherichia coli O157:H7 Sakai genome. PLOS ONE 12: e0184119.
Neuhaus K, Landstorfer R, Fellner L, Simon S, Schafferhans A, Goldberg T, Marx H, Ozoline ON, Rost B, Kuster B, Keim DA and Scherer S (2016) Translatomics combined with transcriptomics and proteomics reveals novel functional, recently evolved orphan genes in Escherichia coli O157:H7 (EHEC). BMC Genomics 17:133.
Fellner L, Simon S, Scherling C, Witting M, Schober S, Polte C, Schmitt-Kopplin P, Keim DA, Scherer S, Neuhaus K (2015) Evidence for the recent origin of a bacterial protein-coding, overlapping orphan gene by evolutionary overprinting. BMC Evolutionary Biology 15:283.
Simon S, Mittelstädt S, Kwon BC, Stoffel A, Landstorfer R, Neuhaus K, Mühlig A, Scherer S, Keim DA (2015) VisExpress: Visual exploration of differential gene expression data. Info Vis 16:48-73.
Fellner L, Bechtel N, Witting MA, Simon S, Schmitt-Kopplin P, Keim D, Scherer S, Neuhaus K (2013) Phenotype of htgA (mbiA), a recently evolved orphan gene of Escherichia and Shigella, completely overlapping in antisense of yaaW. FEMS Microbiol Lett 350:57-64.
Mir K, Neuhaus K, Scherer S, Bossert M, Schober S (2012) Predicting statistical properties of open reading frames in bacterial genomes. PLOS one 7:e45103.
Neuhaus K, Oelke D, Fürst D, Scherer S, Keim DA (2010) Towards automatic detecting of overlapping genes - Clusters BLAST analysis of viral genomes. Pizzuti C, Ritchie MD, Giacobini M (Eds.). EvoBio 2010, LNCS 6023, 228-239.
Loessner M, Gäng S, Scherer S (1999) Evidence for a Holin-Like Protein Gene Fully Embedded Out of Frame in the Endolysin Gene of Staphylococcus aureus Bacteriophage 187. J Bacteriol 181:4452-4460.