Overlapping Genes: Bioinformatics and Evolution
Group leader until 2021: Dr. Zachary Ardern
The redundancy of the genetic code, whereby an amino acid can be encoded by more than one possible nucleotide triplet (codon), enables overlapping protein sequences to be encoded in alternate reading frames of a nucleotide sequence.
The ‘overlapping genes: bioinformatics and evolution group’ is engaged in inferring the presence of unannotated overlapping genes (OLGs) in bacteria using the wealth of existing relevant bioinformatic data. This includes data on transcription and translation from various kinds of RNA sequencing as well as proteomics. Within RNA sequencing we have a particular focus on ribosome profiling.
We are also interested in the evolutionary history of OLGs, and what is required for a successful ‘overprinting’ event leading to an overlapping gene pair. The evolutionary aspect of our research includes investigating the structure of the genetic code and its contribution to overlapping protein-coding sequences.
TUM Masters students interested in conducting practical research or thesis work with us please contact Dr Zachary Ardern.
Publications
Nelson CW, Ardern Z, Goldberg TL, Meng C, Kuo C-H, Ludwig C, Kolokotronis S-O, Wei X (2020) Dynamically evolving novel overlapping gene as a factor in the SARS-CoV-2 pandemic. eLife2020, 9:e59633, Doi:10.7554/eLife.59633
Ardern Z, Neuhaus K, Scherer S (2020) Are antisense proteins in Prokaryotes functional? Frontiers in Molecular Biosciences Vol 7:187. Doi:10.3389/fmolb.2020.00187
Glaub A, Huptas C, Neuhaus K, Ardern Z (2020) Recommendations for bacterial ribosome profiling experiments based on bioinformatics evaluation of published data. Journal of Biological Chemistry 3, 295(27):8999-9011. Doi:10.1074/jbc.RA119.012161
Nelson CW, Ardern Z, Wie X (2020) OLGenie: Estimating Natural Selection to Predict Functional Overlapping Genes. Molecular Biology and Evolution1-9.
Doi:10.1093/molbev/msaa087
Wichmann S, Ardern Z (2019) Optimality in the standard genetic code is robust with respect to comparison code sets. BioSystems 185:104023. Doi:10.1016/j.biosystems.2019.104023
Scherer S, Neuhaus K, Bossert M, Mir K, Keim D, Simon S (2018) Finding new overlapping genes and their theory (FOG Theory). In: Information - and Communication Theory in Molecular Biology, edited by Bossert M (Springer International), pp 137-159.
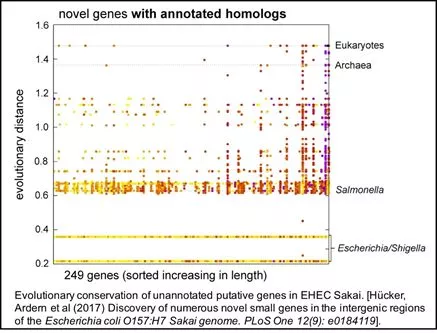
Hücker SM, Ardern Z, Goldberg T, Schafferhans A, Bernhofer M, Vestergaard G, Nelson CW, Schloter M, Rost B, Scherer S, Neuhaus K (2017) Discovery of numerous novel small genes in the intergenic regions of the Escherichia coli O157:H7 Sakai genome. PLoS One. 2017 Sep 13;12(9): e0184119
Mir K, Neuhaus K, Scherer S, Bossert M, Schober S (2012) Predicting statistical properties of open reading frames in bacterial genomes. PLOS one 7, 9,e45103:1-12
Neuhaus K, Oelke D, Fürst D, Scherer S, Keim DA (2010) Towards automatic detecting of overlapping genes - Clusters BLAST analysis of viral genomes. Pizzuti C, Ritchie MD, Giacobini M (Eds.). EvoBio 2010, LNCS 6023, 228-239.